Using Structure to Better Understand Biofilm Formation: An Interview with Lindsey Marmont
Written by Anastassia Pogoutse
A perusal of the “News and Events” section of the Biochemistry website reveals that members of our department regularly produce some very exciting work. However, when a news story distills down a publication to its barest and most interesting facts, it usually washes out any details of the sometimes long and arduous process that led to the breakthrough. Hearing about the experiences of students and postdocs firsthand can provide another story – one filled with failed experiments, sleepless nights, and the feeling of triumph (relief?) at seeing your manuscript finally accepted. Transcripts provides a space for sharing some stories about these behind-the-scenes workings of research.
I recently sat down with Lindsey Marmont, a PhD student in the Howell lab, whose paper was published this past March in PNAS. Lindsey’s research focuses on an exopolysaccharide export system found in the opportunistic pathogen Pseudomonas aeruginosa. The exported molecule, called Pel polysaccharide (PEL), is an essential part of the “glue” that holds P. aeruginosa biofilms together. On top of that, Pel makes the bacteria in the biofilm resistant to treatment with some commonly-used antibiotics. Lindsey’s paper provides insight into how one piece of the Pel export machinery, PelC, guides PEL across the outer membrane on its way out of the cell. The authors used multiple approaches to probe the architecture of the PEL export system, including x-ray crystallography, chemical cross-linking, and biofilm assays. I got the chance to talk to Lindsey about her work and her experiences as a grad student.

Transcripts: Can you talk a little bit about biofilms in Pseudomonas aeruginosa? Why are biofilms such a big issue?
LM: Once the bacteria establish themselves in a biofilm they’re much more difficult to eradicate. Treatments with antibiotics are not as effective, and biofilms are resistant to other things like chemical treatment or the host immune system. It becomes imperative that we know what’s in the biofilm so we can start to target those components and make the bacteria more susceptible to antibiotics.
Transcripts: You went after the PelC protein with the aim of solving a structure. Is there a reason that you went after that particular protein?
LM: In our lab we’re studying a few different exopolysaccharides that are important for biofilm formation in Pseudomonas aeruginosa, and one of those is PEL. PEL is important for the initiation and maintenance of biofilms, and it’s also been shown to increase the tolerance of bacteria to aminoglycoside antibiotics. PelC is actually unique to the Pel system – there is no other polysaccharide system that has a protein similar to PelC and so this presents a unique target.

Transcripts: Was it challenging to solve the structure? Was it hard to get it [PelC] to crystallize?
LM: There were challenges along the way. Someone had started the project before I joined the lab but was unable to solve the P. aeruginosa PelC structure, and so the approach I took was to use PelC orthologs. We found the operon in several other Proteobacteria and I selected a couple of orthologs from that list, and that seemed to do the trick. The orthologs have different surface properties compared to P. aeruginosa PelC, making them more amenable to crystallization.
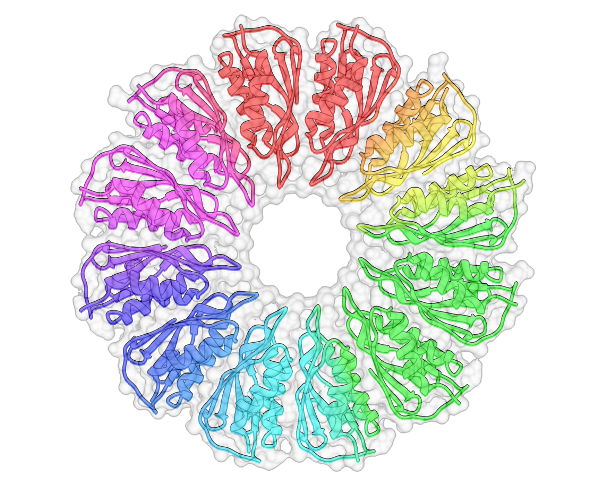
Transcripts: So you solved the structure, and it’s this massive, massive complex. My understanding is that it’s a unique sort of structure?
LM: It’s a really small protein, and it seems to have a fold that’s predisposed to forming oligomers. A lot of structurally similar proteins are involved in multimerization – one I mention in my paper is CsgG and another one that I talk about is FlgT. They all form rings. PelC is unique in some ways but it is that type of fold that also forms a ring.
Transcripts: But there is this negatively charged funnel that you think is important for the Pel…
LM: Alone the monomer doesn’t really tell you much, but when you see the ring as a whole, the concave surface is really negatively charged. By making mutations we found that one [residue] in the centre of the pore, as well as some that are really close to the pore, had an effect on biofilm formation. It suggests that as the positively charged exopolysaccharide is traveling through towards the centre of the pore the negative charge becomes more important – it’s a molecular funnel.
Transcripts: What was the hardest part of that whole project? Because you did some cross-linking, you looked at biofilm formation. What was the hardest experiment?
LM: That’s an interesting question. I think probably the disulfide cross-linking. At least we had the structure before I attempted this so it helped me gauge what residues to try, but I did try a lot of residues to get it to work. There are so many factors going on there, like the proximity of cysteines to each other, and whether the residues I selected to mutate from the structure of Paraburkholderia phytofirmans PelC will be in the exact same position as in P. aeruginosa PelC. But it was exciting when it did work!
Transcripts: Can you use the structure you solved to design some sort of therapeutic? Is there some pharmaceutical application?
LM: The Pel polysaccharide is a good target because it has been shown to be overproduced in sputum isolates (mucous collected from the airways) from cystic fibrosis patients. Since Pel is produced in P. aeruginosa found in cystic fibrosis chronic infections you could design inhibitors targeting that protein, disrupt Pel synthesis and biofilm formation, and in turn render P. aeruginosa more susceptible to antibiotics.
Transcripts: Did you know before you started grad school that you wanted to go into crystallography?
LM: I was part of U Waterloo’s co-op program. I did my 8-month co-op in Lynne’s lab. It gave me a bit of a head start and I got to learn about this subject area, and I really grew to love it. Before that I was already interested in protein structure and trying to figure out how to get from genes to 3D protein structures. Working in the lab really helped solidify that interest, and now my project has even grown into microbiology and doing some bacterial genetics, which I didn’t expect when I started but I really like how it has turned out.
Transcripts: What would be your advice to grad students just starting and looking for a lab?
LM: If you’re interviewing with someone, just ask a lot of questions and figure out what’s important to you. If you want to talk with your PI on a daily basis, or if you want them to give you space. What’s the lab dynamic like? Is there room for the project to expand if you want to do your Ph.D.? Ask all the questions you need to see if that lab is a good fit for you.
Transcripts: What’s next for your project, or are you expanding into something else?
LM: I’m still working on other aspects of the Pel system. I’m looking at other Pel proteins and doing a bit more structure-function and interaction studies to see how all the pieces fit together.

Citation: Lindsey S. Marmont et al. 2017. Oligomeric lipoprotein PelC guides Pel polysaccharide export across the outer membrane of Pseudomonas aeruginosa. PNAS 114 (11): 2892–2897; doi: 10.1073/pnas.1613606114
This interview has been edited for clarity and conciseness, and was view by LM prior to publication. Header illustration by Nikko Torres.
